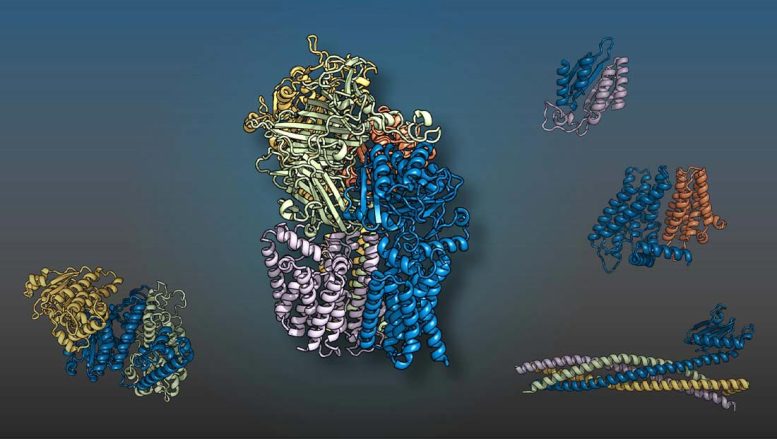
The yeast proteins displayed in various colors come together as 2-, three-, four-, and five-member complexes like 3D puzzle pieces to perform cellular functions. A worldwide group led by scientists at UT Southwestern and the University of Washington predicted the structures using artificial intelligence methods. Credit: UT Southwestern Medical Center
Research led by UT Southwestern and the University of Washington could lead to a wealth of drug targets.
UT Southwestern and University of Washington researchers led a global team that utilized artificial intelligence (AI) and evolutionary analysis to produce 3D designs of eukaryotic protein interactions. The study, released in Science, recognized more than 100 possible protein complexes for the very first time and supplied structural designs for more than 700 previously uncharacterized ones. Insights into the ways sets or groups of proteins fit together to perform cellular processes could result in a wealth of brand-new drug targets.
” Our outcomes represent a considerable advance in the new era in structural biology in which calculation plays a fundamental function,” said Qian Cong, Ph.D., Assistant Professor in the Eugene McDermott Center for Human Growth and Development with a secondary appointment in Biophysics.
Qian Cong, Ph.D. Credit: UT Southwestern Medical
Dr. Cong led the research study with David Baker, Ph.D., Professor of Biochemistry and Dr. Congs postdoctoral coach at the University of Washington prior to her recruitment to UT Southwestern. The study has four co-lead authors, consisting of UT Southwestern Computational Biologist Jimin Pei, Ph.D
. Proteins frequently operate in groups or sets understood as complexes to accomplish every task needed to keep an organism alive, Dr. Cong described.
Till just recently, a major barrier for constructing an interactome was uncertainty over the structures of many proteins, an issue researchers have been trying to fix for half a century. In 2020 and 2021, a business called DeepMind and Dr. Bakers laboratory individually launched two AI technologies called AlphaFold (AF) and RoseTTAFold (RF) that use different strategies to anticipate protein structures based upon the series of the genes that produce them.
In the current study, Dr. Cong, Dr. Baker, and their associates expanded on those AI structure-prediction tools by modeling numerous yeast protein complexes. Yeast is a common design organism for basic biological studies. To discover proteins that were likely to communicate, the scientists first browsed the genomes of associated fungis for genes that acquired anomalies in a linked fashion. They then used the 2 AI innovations to determine whether these proteins could be meshed in 3D structures.
Their work identified 1,505 probable protein complexes. Of these, 699 had actually already been structurally identified, validating the utility of their approach. Nevertheless, there was just minimal experimental information supporting 700 of the predicted interactions, and another 106 had never been described.
To better comprehend these poorly identified or unknown complexes, the University of Washington and UT Southwestern groups worked with colleagues around the globe who were currently studying these or comparable proteins. By integrating the 3D models the researchers in the present research study had generated with information from collaborators, the groups were able to acquire brand-new insights into protein complexes included in maintenance and processing of genetic information, cellular building and construction and transportation systems, metabolism, DNA repair work, and other locations. They likewise determined functions for proteins whose functions were formerly unidentified based on their recently identified interactions with other well-characterized proteins.
” The work described in our brand-new paper sets the phase for similar studies of the human interactome and might ultimately help in establishing new treatments for human illness,” Dr. Cong included.
Dr. Cong noted that the predicted protein complex structures created in this research study are readily available to download from ModelArchive. These others and structures created utilizing this technology in future studies will be a rich source of research concerns for many years to come, she said.
Reference: “Computed structures of core eukaryotic protein complexes” by Ian R. Humphreys, Jimin Pei, Minkyung Baek, Aditya Krishnakumar, Ivan Anishchenko, Sergey Ovchinnikov, Jing Zhang, Travis J. Ness, Sudeep Banjade, Saket R. Bagde, Viktoriya G. Stancheva, Xiao-Han Li, Kaixian Liu, Zhi Zheng, Daniel J. Barrero, Upasana Roy, Jochen Kuper, Israel S. Fernández, Barnabas Szakal, Dana Branzei, Josep Rizo, Caroline Kisker, Eric C. Greene, Sue Biggins, Scott Keeney, Elizabeth A. Miller, J. Christopher Fromme, Tamara L. Hendrickson, Qian Cong and David Baker, 11 November 2021, Science.DOI: 10.1126/ science.abm4805.
Dr. Cong is a Southwestern Medical Foundation Scholar in Biomedical Research. Other UTSW scientists who added to this study consist of Jing Zhang and Josep Rizo, Ph.D., who holds the Virginia Lazenby OHara Chair in Biochemistry.
Working together organizations include: Harvard University, Wayne State University, Cornell University, MRC Laboratory of Molecular Biology, Memorial Sloan Kettering Cancer Center, Gerstner Sloan Kettering Graduate School of Biomedical Sciences, Fred Hutchinson Cancer Research Center, Columbia University, University of Würzburg in Germany, St Jude Childrens Research Hospital, FIRC Institute of Molecular Oncology in Milan, Italy, and the National Research Council, Institute of Molecular Genetics in Rome, Italy..
This work was supported by Southwestern Medical Foundation, the Cancer Prevention and Research Institute of Texas (CPRIT) (RP210041), Amgen, Microsoft, the Washington Research Foundation, Howard Hughes Medical Institute, National Science Foundation (DBI 1937533), National Institutes of Health (R35GM118026, R01CA221858, R35GM136258, R21AI156595), UK Medical Research Council (MRC_UP_1201/ 10), HHMI Gilliam Fellowship, the Deutsche Forschungsgemeinschaft (KI-562/ 11-1, KI-562/ 7-1), AIRC detective and the European Research Council Consolidator (IG23710 and 682190), Defense Threat Reduction Agency (HDTRA1-21-1-0007), and the National Energy Research Scientific Computing Center.
Proteins often run in groups or sets known as complexes to accomplish every job required to keep an organism alive, Dr. Cong explained. In the present research study, Dr. Cong, Dr. Baker, and their coworkers broadened on those AI structure-prediction tools by modeling lots of yeast protein complexes. To better comprehend these badly identified or unidentified complexes, the University of Washington and UT Southwestern groups worked with coworkers around the world who were currently studying these or comparable proteins. By combining the 3D models the researchers in the existing research study had produced with details from collaborators, the groups were able to acquire new insights into protein complexes included in maintenance and processing of genetic details, cellular building and transport systems, metabolic process, DNA repair, and other locations. They likewise identified functions for proteins whose functions were previously unknown based on their freshly recognized interactions with other well-characterized proteins.