Chromosomal analyses have yet to be carried out since the 2 published ctenophore genomes lacked chromosome-level resolution. Prior to diving into the genome, the team of researchers led by Monterey Bay Aquarium Research Institute (MBARI) scientist Darrin Schultz took benefit of the fish tanks capability to culture the California sea gooseberry (Hormiphora californensis) to create the first karyotype for the group. In the case of H. californensis, for instance, a heat map of Hi-C interactions recapitulates the 13 chromosomes visualized microscopically.The group also supplied a preliminary, manually curated annotation for their new genome. A high-quality, chromosome-level genome assembly for the species, released in the November volume of Genome Biology and Evolution, supplies insights that might prove important to resource managers and evolutionary botanists alike. Chickpea (Cicer arietinum) While the first draft of the chickpea genome was published eight years ago, an enormous sequencing effort of more than 3,000 ranges has produced a various genomic first for the species: a pangenome.
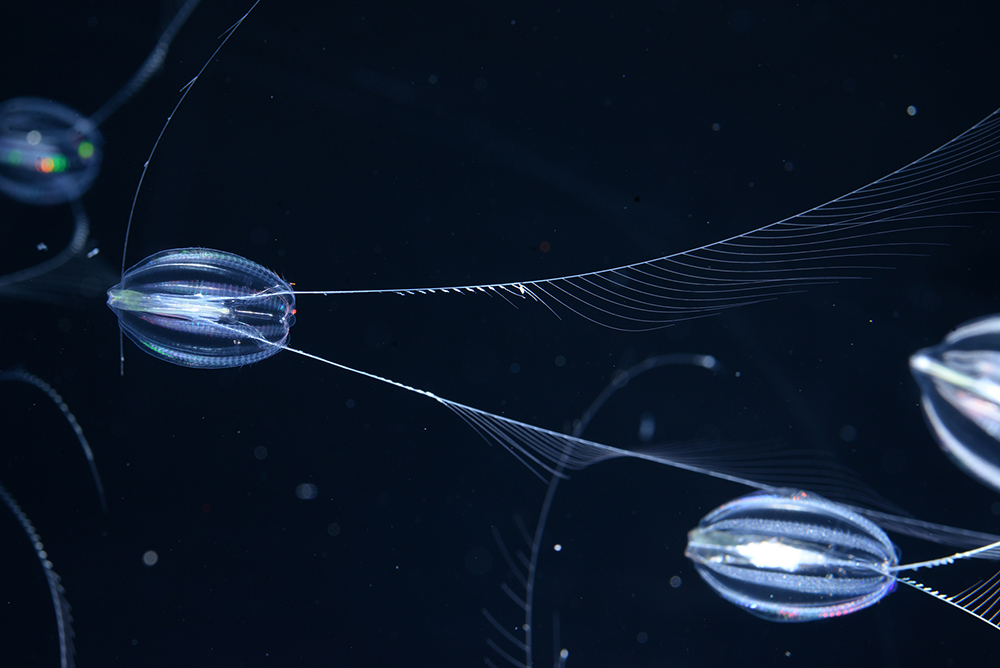
Genetics might offer the opportunity to settle once and for all whether the Porifera or the Ctenophora progressed initially. “One approach that might possibly resolve the phylogenetic position of ctenophores and sponges is comparing whole-chromosomes,” the scientists composed in their paper.With their lack of real tissues and organs and even balance, sponges may look like the obvious choice for the oldest animal group. Beginning back in the early 2010s, a singing group of researchers armed with genomic information started to argue that despite their apparent intricacy, ctenophores (typically referred to as comb jellies) are really more ancient. Proponents of a sponge-first geography clapped back, obviously, and the back-and-forth continues today. Hormiphora californensis is among the types raised for research study and screen by the personnel at the Monterey Bay Aquarium.Darrin Schultz © 2021 MBARIOne possible line of evidence that has remained unexamined is a phylogenomic comparison of whole chromosomes. Chromosomal analyses have yet to be carried out due to the fact that the two published ctenophore genomes lacked chromosome-level resolution. Prior to this research study, it wasnt clear what the chromosomes of ctenophores looked like– there were no released karyotypes for any comb jelly. So before diving into the genome, the team of scientists led by Monterey Bay Aquarium Research Institute (MBARI) scientist Darrin Schultz took advantage of the fish tanks capability to culture the California sea gooseberry (Hormiphora californensis) to produce the first karyotype for the group. The animal, it turns out, has 13 paired chromosomes, which is exactly what was forecasted for a related ctenophore. The team created an initial draft genome from PacBio long reads and polished using Illumina brief paired-end reads. To put the DNA in chromosomal context, the team turned to Hi-C chromatin conformation capture. Hi-C supplies insights into chromatin structure because before series are created, the native DNA is treated with formaldehyde, which generates crosslinks between engaging pieces of chromatin. When the DNA is digested, these crosslinks initially remain intact, keeping together pieces of sequence that physically connect (primarily within chromosomes). Attached sequences are then fused to one another, producing chimeric series that maintain the genomic spatial context of undamaged chromosomes. In the case of H. californensis, for example, a heat map of Hi-C interactions recapitulates the 13 chromosomes visualized microscopically.The group also supplied a preliminary, manually curated annotation for their new genome. This exposed thousands of nested intronic genes: genes that exist within the introns of other genes. In fact, 484 were twice as nested. Research suggests that nesting may permit for more fine-tuned transcription control, and for that reason might have played a crucial function in the evolution of intricacy. Indeed, such nesting of genes is seen in plants and in animals, consisting of in humans, embedded intronic genes are largely missing from fungis and single-celled eukaryotes. The authors keep in mind that finding many of these genes within genes in this ctenophore– on par with the degree of nesting seen in primates– may recommend the mechanism developed early on in our lineage.While the researchers didnt run whole-chromosome-level phylogenomic analyses to see if the H. californensis genome contributes to the support for a ctenophore-first evolutionary history, such analyses are now possible, as a chromosome-level assembly for a sponge species was released last year. Runners Up: Japanese stiltgrass (Microstegium vimineum) Japanese stiltgrass has actually spent more than a century wreaking havoc on the native environments of the eastern US, yet the biological basis of its invasiveness has stayed elusive. A high-quality, chromosome-level genome assembly for the types, published in the November volume of Genome Biology and Evolution, offers insights that could show important to resource supervisors and evolutionary botanists alike. Chickpea (Cicer arietinum) While the first draft of the chickpea genome was released eight years back, a huge sequencing effort of more than 3,000 varieties has produced a various genomic first for the species: a pangenome. As The New York Times describes, this brochure of intraspecies hereditary variation sheds light on the origins and domestication history of this crucial crop, and might assist farming researchers breed or engineer new ranges that are able to endure whatever environment change throws their way. See “The Pangenome: Are Single Reference Genomes Dead?”.