Did modern-day mammal groups co-exist with the dinosaurs, or did they come from after the mass extinction?” The timeline of mammal evolution is possibly one of the most controversial topics in evolutionary biology. In our case, this was hard due to the gigantic dataset examined, involving hereditary data from practically 5,000 mammal species and 72 complete genomes,” Dr. dos Reis states.
“We fixed the computational obstacles by dividing the analysis in sub-steps: first simulating timelines using the 72 genomes and then utilizing the outcomes to guide the simulations on the staying types. “If we had attempted to analyze this large mammal dataset in a supercomputer without using the Bayesian technique we have actually established, we would have had to wait years to infer the mammal timetree.
The research group was led by Dr. Mario dos Reis (Queen Mary University of London) and Professor Phil Donoghue (University of Bristol), and consisted of scientists from Queen Mary, University of Bristol, UCL, Imperial College London, and the University of Cambridge.
Dr. Sandra Álvarez-Carretero, lead author of the paper from UCL (then at Queen Mary), says: “By integrating complete genomes in the analysis and the needed fossil details, we had the ability to reduce uncertainties and obtain an exact evolutionary timeline. Did modern mammal groups co-exist with the dinosaurs, or did they originate after the mass termination? We now have a certain response.”
” The timeline of mammal development is possibly one of the most controversial topics in evolutionary biology. Early studies supplied origination estimates for modern-day placental groups deep in the Cretaceous, in the dinosaur age. The previous 20 years had actually seen studies moving back and forth in between post- and pre-K-Pg diversification situations. Our precise timeline settles the concern,” includes Prof Donoghue, co-senior author of the paper.
Quick method for genome analysis
With around the world sequencing tasks now producing hundreds to countless genome sequences, and with imminent plans to series more than a million species, evolutionary biologists will quickly have a wealth of info at their hands. Present methods to analyze the large genomic datasets readily available and create evolutionary timelines are ineffective and computationally pricey.
” Inferring evolutionary timelines is an essential objective of biology. Cutting edge techniques rely on utilizing computers to replicate evolutionary timelines and evaluate the most possible ones. In our case, this was hard due to the gigantic dataset examined, including hereditary data from almost 5,000 mammal types and 72 total genomes,” Dr. dos Reis says.
In this study, the scientists established a new, fast Bayesian approach to examine big numbers of genome series, whilst also representing uncertainties within the information. “We solved the computational difficulties by dividing the analysis in sub-steps: first replicating timelines utilizing the 72 genomes and then utilizing the results to direct the simulations on the remaining types. Using genomes minimizes unpredictability because it permits rejection of unplausible timelines from the simulations,” says Dr dos Reis.
” Our information processing pipeline sourced as much genomic data for as lots of mammal types as possible. This was challenging because hereditary databases contain inaccuracies and we needed to establish a method to determine poor quality samples or mislabelled data that had actually to be eliminated,” adds Dr. Asif Tamuri, co-lead author of the paper from UCL, who was responsible for assembling the mammal genomic dataset.
More efficient and sustainable
Using their unique method, the group was able to decrease calculation time for this complex analysis from decades to months. “If we had attempted to analyze this large mammal dataset in a supercomputer without utilizing the Bayesian technique we have actually developed, we would have had to wait decades to infer the mammal timetree.
The method developed in the research study could be utilized to take on other contentious evolutionary timelines that need analysis of big datasets. By integrating the unique Bayesian approach with the forthcoming genomes from the Darwin Tree of Life and Earth BioGenome tasks, the idea of approximating a trusted evolutionary timescale for the Tree of Life now appears within reach.
Recommendation: “A Species-Level Timeline of Mammal Evolution Integrating Phylogenomic Data” by Sandra Álvarez-Carretero, Asif U. Tamuri, Matteo Battini, Fabrícia F. Nascimento, Emily Carlisle, Robert J. Asher, Ziheng Yang, Philip C. J. Donoghue and Mario dos Reis, 22 December 2021, Nature.DOI: 10.1038/ s41586-021-04341-1.
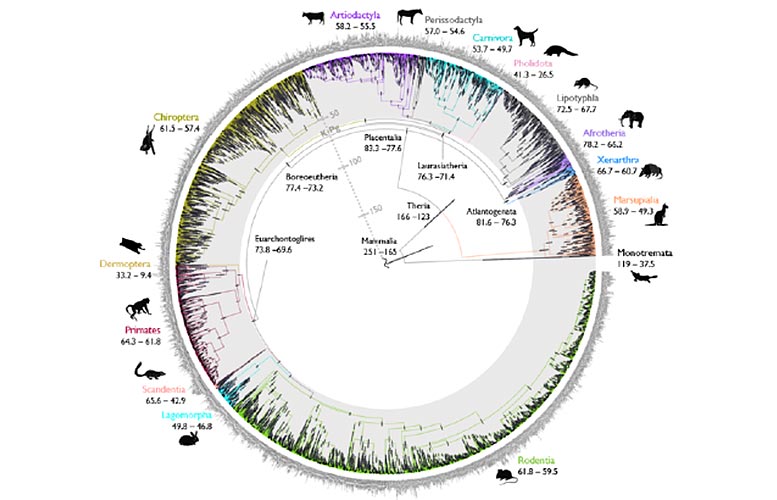
Mammal tree of life. Credit: Mario dos Reis Barros and Sandra Alvarez-Carretero
A brand-new research study, published on December 22, 2021, in the journal Nature, has actually offered the most comprehensive timeline of mammal advancement to date.
The research describes a quick and new computational approach to acquire specifically dated evolutionary trees, referred to as timetrees. The authors utilized the unique approach to analyze a mammal genomic dataset and answer an enduring concern around whether contemporary placental mammal groups originated prior to or after the Cretaceous-Palaeogene (K-Pg) mass extinction, which cleaned out over 70 percent of all species, including all dinosaurs.
The findings validate the forefathers of modern placental mammal groups postdate the K-Pg termination that happened 66 million years ago, settling a controversy around the origins of contemporary mammals. Placental mammals are the most varied group of living mammals, and include groups such as primates, rodents, cetaceans, carnivorans, chiropterans (bats) as well as human beings.