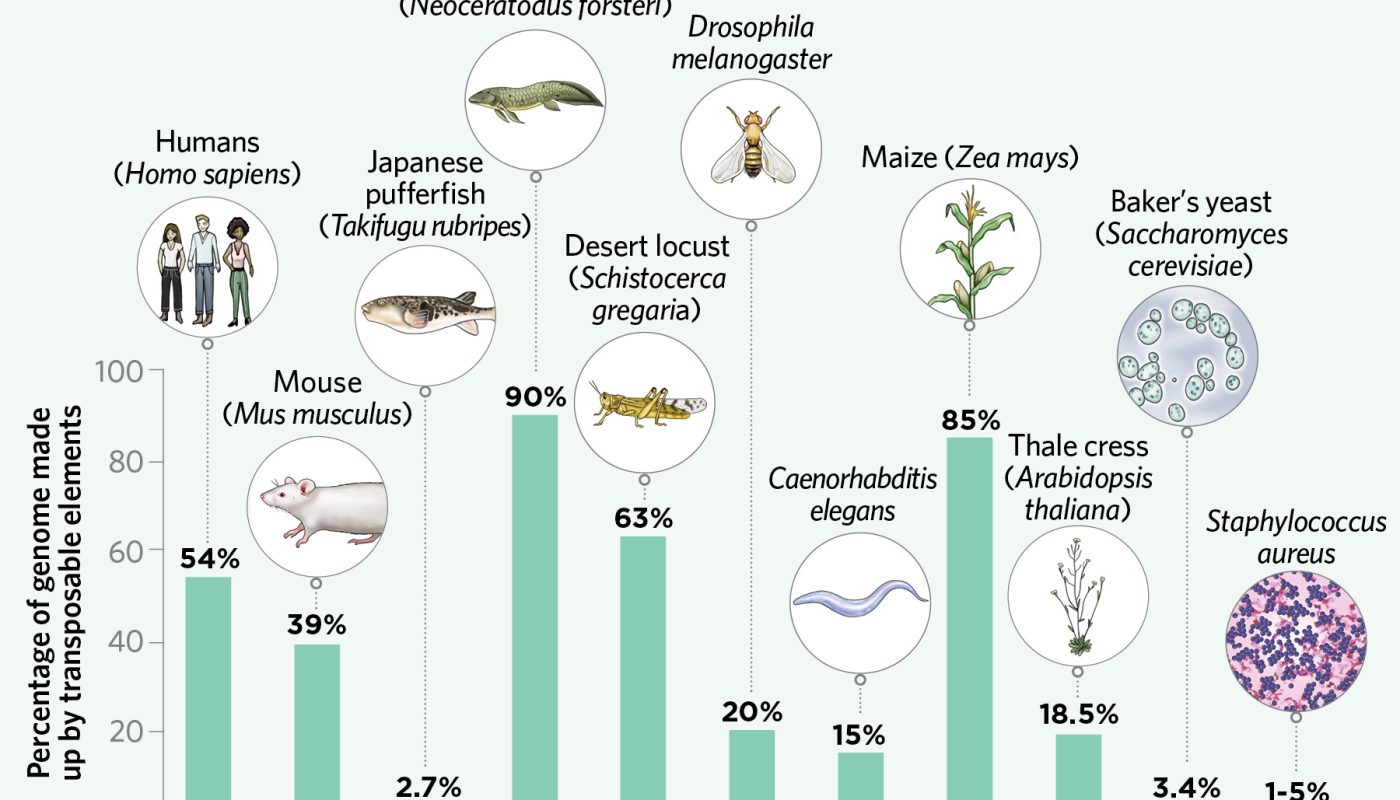
There are various manner ins which mobile genetic aspects can affect development. Numerous transposable aspects (TEs), frequently called transposons, contain genes that code for their jumping or copying equipment, and over time these may be “domesticated” through anomaly and selection, ending up being integral parts of the organisms genome. The RAG1 and RAG2 enzymes that mix up DNA sectors in immune proteins (antibodies and T cell receptors) are a noteworthy example. “Wild” TEs can likewise have adaptive capacity, producing genetic diversity as they leap. If TEs land inside a gene, they can directly change coding regions, mRNA splice websites, or expression-related motifs. And due to the fact that transposons frequently consist of transcription aspect binding sites and other regulative sequences, they can change a genes expression even if they land close by. The transposable aspects can likewise modify the genome in other methods– such as by selecting up substantial pieces of DNA as they jump– that scientists suspect are similarly changing the course of evolution.Inserting into GenesArguably the most immediate and dramatic impacts TEs have on genomes occur when they place into active genes. They can jump into coding areas, altering protein sequences, or they can insert into noncoding regions and alter gene splicing or expression. This is what took place in peppered moths, when a 22-kb TE placed into the cortex gene and resulted in overproduction of melanin, turning dark the generally lightly bespeckled moths and enhancing their survival in contaminated environments.Inserting near GenesUnlike point anomalies, someTEs come preloaded with genetic concepts that might impact the expression of nearby genes. Specific populations of Drosophila carry the TE insertion FBti0019386, for example, which consists of transcription element (TF) binding sites that are activated during a bacterial infection which boost expression of the immune-related gene Bin1. Flies carrying FBti0019386 are most likely to endure shot with a pathogenic pressure of Pseudomonas.VARIABLE ELEMENTS “You can discover transposable elements in virtually all the organisms that have been studied [genetically], from germs to eukaryotes,” notes genomicist Josefa González Pérez of Pompeu Fabra University in Barcelona. But while TEs are nearly universal throughout living organisms, their frequency varies extensively. In some organisms, TEs dominate, representing approximately 90 percent of the genome, while in others, transposable elements comprise just a fraction of the whole genetic code. When plentiful, TEs can grow the size of the genome to huge, unwieldy percentages that continue to baffle scientists.Data from: PLOS Genet, 17: e1009768, 2021; Plant Physiol, 139:1612– 24, 2005; Genome Biol Evol, 5:1886– 901, 2013; Genome Biol, 10:107, 2009; Nature, 590:284– 89, 2021; Science, 297:1301– 10, 2002; Cytogenet Genome Res, 147:217– 39, 2015; bioRxiv, doi:10.1101/ 2021.07.12.451456, 2021; Nature, 420:520– 62, 2002; F1000Res, 9:775, 2020; Mobile DNA, 11:23, 2020; PLOS ONE, 6: e16526, 2011; C. elegans II. Second edition, CSHL Press, 1997; BMC Bioinformat, 20:484, 2019; PLOS ONE, 7: e50978, 2012; Curr Microbiol, 62:198– 208, 2011; J Bacteriol, 194:4124, 2012Read the full story.
Numerous transposable aspects (TEs), often called transposons, include genes that code for their copying or jumping machinery, and over time these might be “domesticated” through mutation and choice, becoming integral parts of the organisms genome. The transposable aspects can also alter the genome in other methods– such as by picking up substantial chunks of DNA as they jump– that researchers think are likewise altering the course of evolution.Inserting into GenesArguably the most immediate and dramatic effects TEs have on genomes occur when they place into active genes. When abundant, TEs can grow the size of the genome to huge, unwieldy proportions that continue to baffle scientists.Data from: PLOS Genet, 17: e1009768, 2021; Plant Physiol, 139:1612– 24, 2005; Genome Biol Evol, 5:1886– 901, 2013; Genome Biol, 10:107, 2009; Nature, 590:284– 89, 2021; Science, 297:1301– 10, 2002; Cytogenet Genome Res, 147:217– 39, 2015; bioRxiv, doi:10.1101/ 2021.07.12.451456, 2021; Nature, 420:520– 62, 2002; F1000Res, 9:775, 2020; Mobile DNA, 11:23, 2020; PLOS ONE, 6: e16526, 2011; C. elegans II.