Scientists at the Icahn School of Medicine at Mount Sinai discovered that the majority of disease-causing anomalies have a low threat of really causing illness. Credit: Courtesy of Do lab, Mount Sinai, N.Y., N.Y.
Results of big biobank research study by Mount Sinai researchers may help doctors better evaluate real illness danger.
Picture getting a positive outcome on a hereditary test. The medical professional informs you that you have a “pathogenic hereditary variant,” or a DNA series that is understood to raise the chances for getting a disease like breast cancer or diabetes. But what exactly are those chances– 10 percent? Fifty percent? One hundred? Currently, that is not a simple concern to respond to.
To resolve this need, researchers at the Icahn School of Medicine at Mount Sinai examined the DNA sequences and electronic health record data of countless individuals saved in 2 massive biobanks. In general, they found that the opportunity a pathogenic genetic variant might in fact cause an illness is fairly low– about 7 percent. Nonetheless, they likewise discovered that some variants, such as those related to breast cancer, are connected to a large range of risks for disease. The results, released in JAMA, could alter the way the threats associated with these variants are reported, and one day, help assist the way physicians translate hereditary screening outcomes.
” A major goal of this study was to produce handy, advanced stats which quantitatively assess the impact that known disease-causing genetic variations may have on an individuals risk to illness,” stated Ron Do, PhD, Associate Professor of Genetics and Genomic Sciences and a member of The Charles Bronfman Institute for Personalized Medicine at Icahn Mount Sinai.
Due to the nature of these discoveries, it has been hard to estimate– or supply stats on– the real risk of this taking place for each gene variant. Studies like these that do not use randomly selected large populations may produce overestimates of the risk presented by variants.
In this study, the scientists tackled the problem by browsing massive DNA sequencing information of 72,434 people for 37,780 recognized versions and then scanning each individuals health records for a corresponding illness medical diagnosis. The comprehensive search included 29,039 individuals in Mount Sinais BioMe ® Biobank program and 43,395 individuals who belonged to the UK Biobank.
The study was led by Iain S. Forrest, an MD-PhD candidate in Dr. Dos lab who found motivation from previous clinical experience he had as part of a postbaccalaureate fellowship at the National Institutes of Health (NIH).
” The idea for the research study came out of a conceptualizing session,” stated Mr. Forrest. “Dr. Do and I talked about the requirement to have a much better system for classifying illness risk. Presently, variants are classified by broad labels such as pathogenic or benign. As I learned in the clinic, theres a lot of grey area with these labels. Thats when we understood that the biobanks which connect DNA series data to electronic health records are an exceptional opportunity to resolve this requirement.”
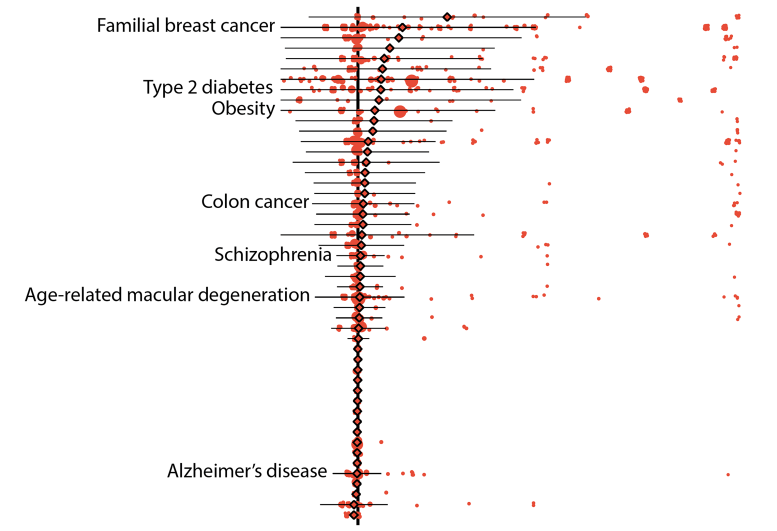
Preliminary results revealed that 157 illness in their information set could be linked to 5,360 versions that were specified as either “pathogenic” by ClinVar, an extensively referenced, NIH-supported public library, or “loss-of-function” as anticipated by bioinformatic algorithms. Typically, the “penetrance,” or opportunity that a variation was linked to a disease medical diagnosis, was low, particularly 6.9 percent. Likewise, the typical risk distinction, which explains the increase in disease danger for an individual who has the version over an individual who does not have it, was also low.
” At first I was rather surprised by the results. The dangers we discovered were lower than I anticipated,” stated Dr. Do. “These outcomes raise questions about how we must be categorizing the risks of these variants.”
Regardless of these results, the risks related to some genetic variants stayed high. Pathogenic variants of the breast cancer genes BRCA1 and BRCA2 both averaged 38 percent penetrance, with private variations falling between zero and 100 percent.
More outcomes demonstrated other benefits of using biobank data. In one example, the researchers had the ability to determine the dangers of individual versions that are associated with age-related disorders, such as some forms of type 2 diabetes and breast and prostate cancers. Usually, the penetrance of these versions had to do with 10 percent for individuals over 70 years of age whereas it had to do with 8 percent for those who were older than 20.
The team likewise found that the presence of some versions could depend on an individuals ethnic culture and determined more than 100 variations that are particularly found in people of non-European descent.
Finally, the authors listed numerous possible methods the study itself might have under- or overestimated the dangers reported.
” While more research is required to be done, we feel that this research study is an excellent very first action towards ultimately supplying medical professionals and clients with the precise and nuanced information they need to make more exact diagnoses,” said Dr. Do.
Reference: “Population-Based Penetrance of Deleterious Clinical Variants” by Iain S. Forrest, BS; Kumardeep Chaudhary, PhD; Ha My T. Vy, PhD; Ben O. Petrazzini, BS; Shantanu Bafna, MS; Daniel M. Jordan, PhD; Ghislain Rocheleau, PhD; Ruth J. F. Loos, PhD; Girish N. Nadkarni, MD; Judy H. Cho, MD and Ron Do, PhD, 25 January 2022, JAMA.DOI: 10.1001/ jama.2021.23686.
This work was supported by the National Institutes of Health (GM124836, GM007280, HL139865, and HL155915).
Research studies like these that do not utilize randomly selected big populations might produce overestimates of the risk posed by versions.
The average threat difference, which explains the boost in illness risk for a person who has the variant over a person who does not have it, was likewise low.
In one example, the researchers were able to compute the risks of specific variations that are associated with age-related disorders, such as some kinds of type 2 diabetes and breast and prostate cancers.
They also discovered that some versions, such as those associated with breast cancer, are linked to a large variety of risks for disease. The results, published in JAMA, could modify the method the risks associated with these variants are reported, and one day, help guide the method physicians analyze hereditary screening results.