” 3D structures of proteins provide us with atomic-resolution information on the structure of SARS-CoV-2 that is vital for developing treatments or vaccines targeting distinct parts of the virus. Thanks to a current research focus on SARS-CoV-2, researchers have actually figured out around a thousand 3D structures of the viruss 27 specific proteins, and almost a thousand more for related proteins,” discusses Professor ODonoghue. The groups analysis exposes opportunities for additional research study. “Much of the coronavirus research study to date has actually focused on the spike glycoprotein, which is the main target for present vaccines. This protein will continue to be an important target, however its likewise important we broaden our focus to other possible targets and better comprehend the whole viral lifecycle,” states Professor ODonoghue.
The landing page of the COVID-19 protein modeling resource in Aquaria. Credit: Garvan Institute of Medical Research
The most comprehensive analysis of the 3D structure of SARS-CoV-2 to date has actually exposed new insight on how the infection duplicates and infects human cells.
Led by Professor Sean ODonoghue, from the Garvan Institute of Medical Research and CSIROs Data61, researchers put together more than 2000 different structures involving the coronaviruss 27 proteins. The analysis identified viral proteins that mimic and hijack human proteins– tactics that permit the virus to bypass cell defenses and reproduce.
These structural models can be freely accessed from the Aquaria-COVID resource, a website developed by the team to help the research study community focus on prospective new targets on the virus for future treatments or vaccines, and most importantly examine brand-new infection versions.
” Our resource includes a level of information of SARS-CoV-2s structure that is not available anywhere else. This has actually provided us an unprecedented insight into the infections activity,” says Professor ODonoghue, first author of a paper in the journal Molecular Systems Biology detailing the groups findings.
The SARS-CoV-2 envelope designed in Aquaria. Credit: Garvan Institute of Medical Research
” Our analysis has highlighted key systems used by the coronavirus; these mechanisms, in turn, may guide the advancement of new therapies and vaccines.”
Structural insights
To better comprehend biological procedures, scientists determine the 3D shape of specific proteins– the building blocks that comprise viruses or cells.
” 3D structures of proteins supply us with atomic-resolution information on the composition of SARS-CoV-2 that is important for establishing vaccines or treatments targeting unique parts of the infection. Thanks to a recent research study concentrate on SARS-CoV-2, scientists have determined around a thousand 3D structures of the infections 27 private proteins, and nearly a thousand more for related proteins,” describes Professor ODonoghue. “However, until now there has been no easy method to bring all the pieces of data together and analyze them.”.
SARS-CoV-2 RNA synthesis complex modeled in Aquaria. Credit: Garvan Institute of Medical Research.
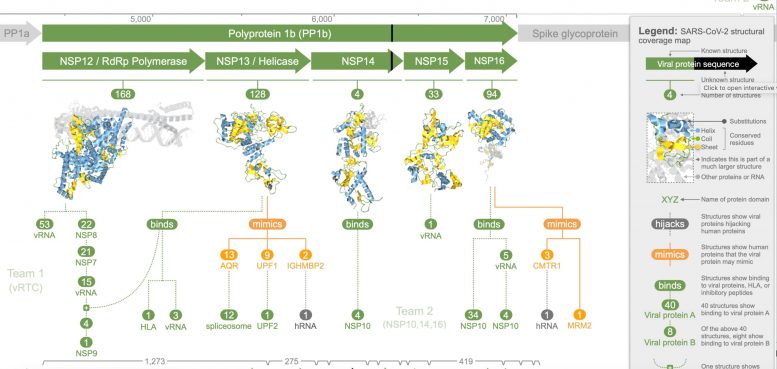
The groups analysis revealed 3 coronavirus proteins (NSP13, nsp3, and nsp16) that imitated human proteins, which the researchers think allows the infection to better conceal from the human body immune system and may add to the variation in COVID-19 outcomes.
The modeling also revealed five coronavirus proteins (NSP1, NSP3, spike glycoprotein, envelope protein, and ORF9b protein) that the researchers say hijack or disrupt procedures in human cells, thus helping the virus take control, complete its life cycle, and spread to other cells.
” Further, we found 8 coronavirus proteins that self-assemble with each other– examining how they assembled has supplied new insights into how the infection reproduces its genome. Nevertheless, after representing overlaps, this still leaves 14 proteins that we believe play essential roles in infection however have no structural evidence of interaction with other viral or human proteins,” states Professor ODonoghue.
SARS-CoV-2 spike glycoprotein and ACE2 protein modeled in Aquaria. Credit: Garvan Institute of Medical Research.
” To make all these insights and data more accessible to scientists, we devised a new visualization method called a structural protection map. The map highlights what we understand about SARS-CoV-2 and what is still delegated discover– it also helps scientists utilize and discover 3D models to investigate particular research study concerns.”.
Viral security.
The groups analysis reveals chances for further research. “Much of the coronavirus research study to date has actually focused on the spike glycoprotein, which is the main target for present vaccines. This protein will continue to be an essential target, but its likewise important we expand our focus to other prospective targets and better comprehend the entire viral lifecycle,” states Professor ODonoghue.
He adds that the Aquaria-COVID resource might help scientists more quickly examine how new variations of coronavirus vary– and critically, how they might better be targeted with treatments and vaccines.
” The longer the infection circulates, the more opportunities it needs to mutate and form brand-new variations such as the Delta strain,” says Professor ODonoghue. “Our resource will assist scientists comprehend how brand-new pressures of the infection differ from each other– a piece of the puzzle that we hope will assist in handling new variants as they emerge.”.
Referral: “SARS-CoV-2 structural protection map reveals viral protein assembly, hijacking, and mimicry systems” 14 September 2021, Molecular Systems Biology.DOI: 10.15252/ msb.202010079.
This research was supported by the Sony Foundation Australia, Tour de Cure Australia, the Wellcome Trust, Biotechnology and Biological Sciences Research Council and the Bundesministerium für Bildung und Forschung (BMBF).
This project was a partnership in between the Garvan Institute of Medical Research, CSIRO Data61, UNSW Sydney, Weihenstephan-Triesdorf University of Applied Sciences, Technical University of Munich, The University of Dundee, and University College London.
Teacher ODonoghue is a Conjoint Professor, School of Biotechnology and Biomolecular Sciences (BABS), UNSW Sydney and a Visiting Scientist, Commonwealth Scientific and Industrial Research Organisation (CSIRO) Data61, Australias nationwide science company.