
Researchers from the Max Planck Institute in Marburg and the Philipps University in Marburg have discovered the first regular molecular fractal in nature. It is an enzyme in cyanobacteria, known as citrate synthase, which assembles into a Sierpiński triangle pattern. This finding was highly unexpected and suggests that nature might hold more intricate designs at the molecular level than previously understood.
Emergence of fractal geometry at the molecule level
Fractals are patterns that repeat themselves at different sizes, creating complex structures from simple rules. They are found everywhere in nature, from the shapes of mountains and coastlines to the branching of trees and veins in leaves. Unlike traditional geometric shapes, fractals can have infinite detail, appearing the same at any scale. This means that whether you zoom in or out, the pattern remains consistent.
However, such regularity has eluded the molecular world until now. Although molecules can self-assemble themselves into all kinds of interesting shapes, they tend to lose their distinct features when viewed en masse, blending into a smooth continuum. This makes the discovery of a regular molecular fractal both surprising and significant.
The research team stumbled upon this molecular fractal while studying microbial enzymes through electron microscopy. The researchers shared their astonishment at the sight of the enzyme’s spontaneous assembly into the iconic fractal pattern of the Sierpiński triangle. The Sierpiński Triangle is created by repeatedly removing a triangle the middle of from a larger triangle, resulting in a pattern of infinitely repeating triangles within triangles.
“We stumbled on this structure completely by accident and almost couldn’t believe what we saw when we first took images of it using an electron microscope” says first author Franziska Sendker of the Max Planck Institute for Terrestrial Microbiology.
“The protein makes these beautiful triangles and as the fractal grows, we see these larger and larger triangular voids in the middle of them, which is totally unlike any protein assembly we’ve ever seen before,” she continues.

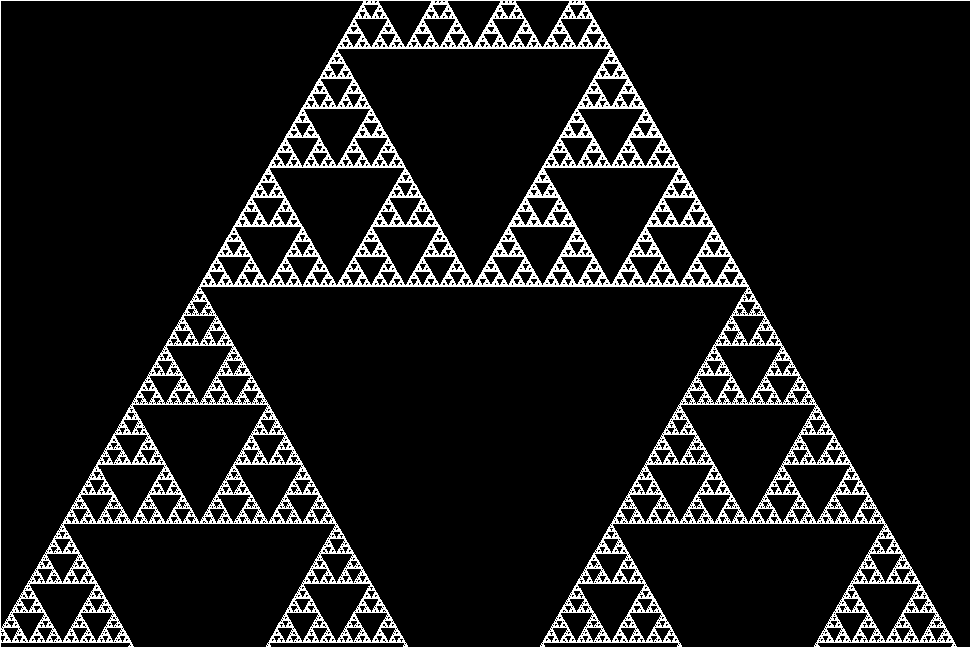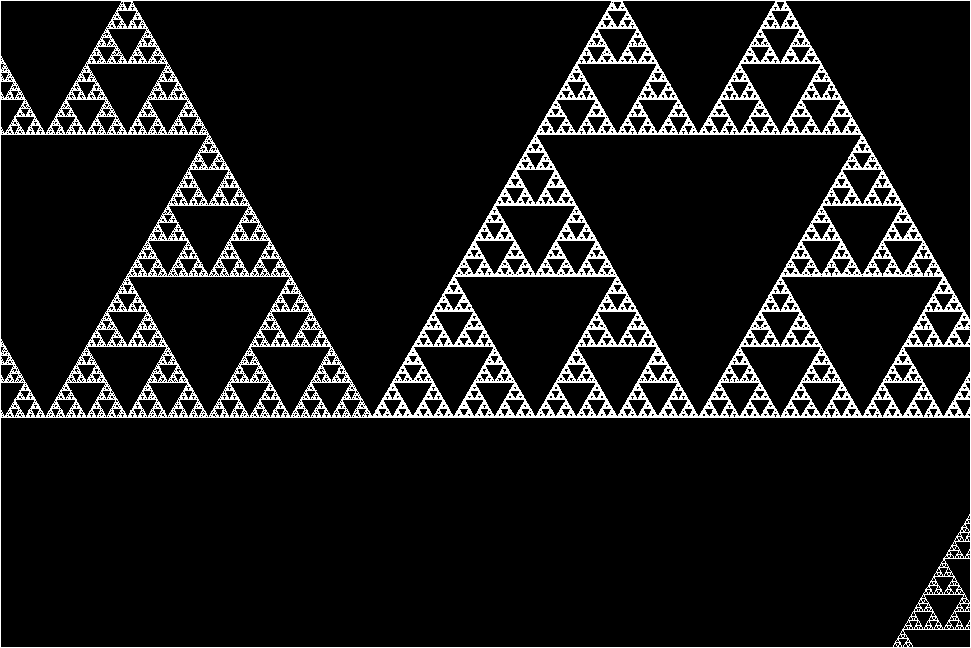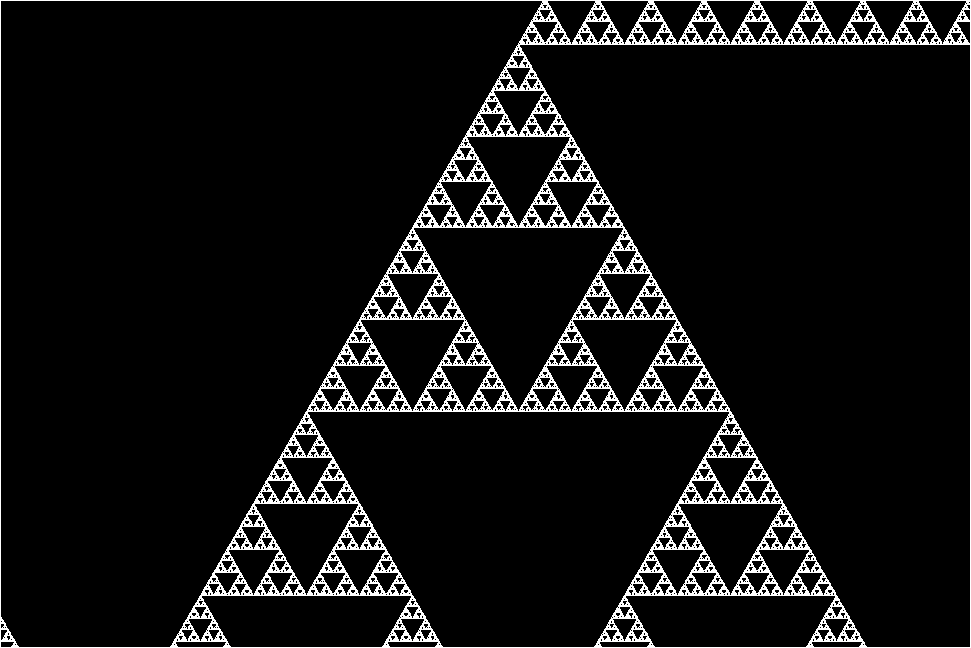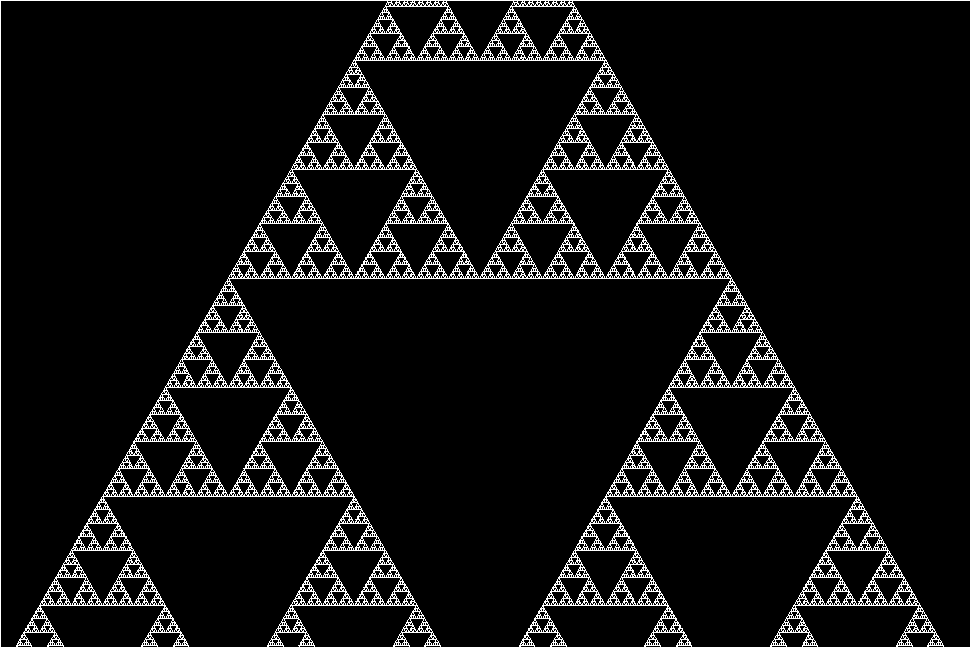
Cracking the fractal code
Understanding how this enzyme achieves its fractal geometry posed a unique challenge. Jan Schuller and his team at the University of Marburg were up to tackle it. They used electron microscopy to unravel the molecular structure of the enzyme assembly. However, the computer that interpreted the data was getting confused by all the smaller triangles that formed the larger ones. The algorithm kept focusing on these smaller units rather than zooming out to see the big picture, missing the forest for the trees.
However, the researchers plowed through and managed to image the enzyme’s structure. They discovered that the key to its fractal formation lies in violating conventional symmetry seen in protein self-assembly. In a typical protein chain, each individual protein adopts the same arrangement relative to its neighbor. But instead of adopting a uniform arrangement, different protein chains interacted in slightly varied ways across the fractal, leading to the emergence of the Sierpiński pattern.
Interestingly, this unique fractal assembly seems completely coincidental — a serendipitous byproduct of evolution. Experiments in which the scientists genetically removed the ability of the citrate synthase to assemble into fractals suggested it doesn’t affect the cyanobacterium’s growth. This means that this complex structure might have arisen from an evolutionary fluke rather than serving a specific biological purpose.
“This prompted us to wonder whether this might just be a harmless accident of evolution. Such accidents can happen when the structure in question isn’t too difficult to construct,” said evolutionary biologist Georg Hochberg, one of the senior authors of the study.
Reverse evolution
To further investigate the origins of the fractal, the researchers reverse-engineered the protein sequence of the fractal enzyme over millions of years. By back-calculating the protein sequence of ancient cyanobacteria and reproducing these proteins, the researchers discovered that the fractal arrangement emerged abruptly from a small number of mutations and was subsequently lost in several lineages of cyanobacteria, surviving in only one species.
There was no pressing evolutionary factor, no good reason for this fractal enzyme to exist. And yet it does. This may mean that there may be other natural fractal proteins waiting to be discovered in the natural world.
The findings appeared in the journal Nature.
Thanks for your feedback!
